Get biological occurrences
Use the 1 million records subsampled from the full OBIS dataset otherwise available at https://obis.org/data/access.
occ <- occ_1M # occ_1M OR occ_SAtlanticCreate a discrete global grid
Create an ISEA discrete global grid of resolution 9 using the dggridR package:
library(dggridR) # remotes::install_github("r-barnes/dggridR")
dggs <- dgconstruct(projection = "ISEA", topology = "HEXAGON", res = 9)Here’s on overview of all possible resolutions specified by the res parameter above:
inf <- dginfo(dggs)
#> res cells area_km spacing_km cls_km
#> 1 0 12 51006562.1724088639021 7053.6524314108 8199.5003701020
#> 2 1 32 17002187.3908029571176 4072.4281300451 4678.9698717297
#> 3 2 92 5667395.7969343187287 2351.2174771369 2691.2520709129
#> 4 3 272 1889131.9323114394210 1357.4760433484 1551.8675487723
#> 5 4 812 629710.6441038132180 783.7391590456 895.6018416484
#> 6 5 2432 209903.5480346043769 452.4920144495 517.0049969031
#> 7 6 7292 69967.8493448681256 261.2463863485 298.4793231872
#> 8 7 21872 23322.6164482893764 150.8306714832 172.3244908961
#> 9 8 65612 7774.2054827631255 87.0821287828 99.4910857272
#> 10 9 196832 2591.4018275877083 50.2768904944 57.4411078487
#> 11 10 590492 863.8006091959028 29.0273762609 33.1636203580
#> 12 11 1771472 287.9335363986343 16.7589634981 19.1470215381
#> 13 12 5314412 95.9778454662114 9.6757920870 11.0545373459
#> 14 13 15943232 31.9926151554038 5.5863211660 6.3823399790
#> 15 14 47829692 10.6642050518013 3.2252640290 3.6848456792
#> 16 15 143489072 3.5547350172671 1.8621070553 2.1274466399
#> 17 16 430467212 1.1849116724224 1.0750880097 1.2282818893
#> 18 17 1291401632 0.3949705574741 0.6207023518 0.7091488792
#> 19 18 3874204892 0.1316568524914 0.3583626699 0.4094272963
#> 20 19 11622614672 0.0438856174971 0.2069007839 0.2363829597
#> 21 20 34867844012 0.0146285391657 0.1194542233 0.1364757654
#> 22 21 104603532032 0.0048761797219 0.0689669280 0.0787943199
#> 23 22 313810596092 0.0016253932406 0.0398180744 0.0454919218
#> 24 23 941431788272 0.0005417977469 0.0229889760 0.0262647733
#> 25 24 2824295364812 0.0001805992490 0.0132726915 0.0151639739
#> 26 25 8472886094432 0.0000601997497 0.0076629920 0.0087549244
#> 27 26 25418658283292 0.0000200665832 0.0044242305 0.0050546580
#> 28 27 76255974849872 0.0000066888611 0.0025543307 0.0029183081
#> 29 28 228767924549612 0.0000022296204 0.0014747435 0.0016848860
#> 30 29 686303773648832 0.0000007432068 0.0008514436 0.0009727694
#> 31 30 2058911320946492 0.0000002477356 0.0004915812 0.0005616287Then assign cell numbers to the occurrence data:
occ$cell <- dgGEO_to_SEQNUM(dggs, occ$decimalLongitude, occ$decimalLatitude)[["seqnum"]]Calculate indicators
The following function calculates the number of records, species richness, Simpson index, Shannon index, Hurlbert index (n = 50), and Hill numbers for each cell.
Perform the calculation on species level data:
idx <- calc_indicators(occ)Add cell geometries to the indicators table (idx):
library(sf)
grid <- dgcellstogrid(dggs, idx$cell) %>%
st_wrap_dateline() %>%
rename(cell = seqnum) %>%
left_join(
idx,
by = "cell")Plot maps of indicators
Let’s look at the resulting indicators in map form.
# ES(50)
gmap_indicator(grid, "es", label = "ES(50)")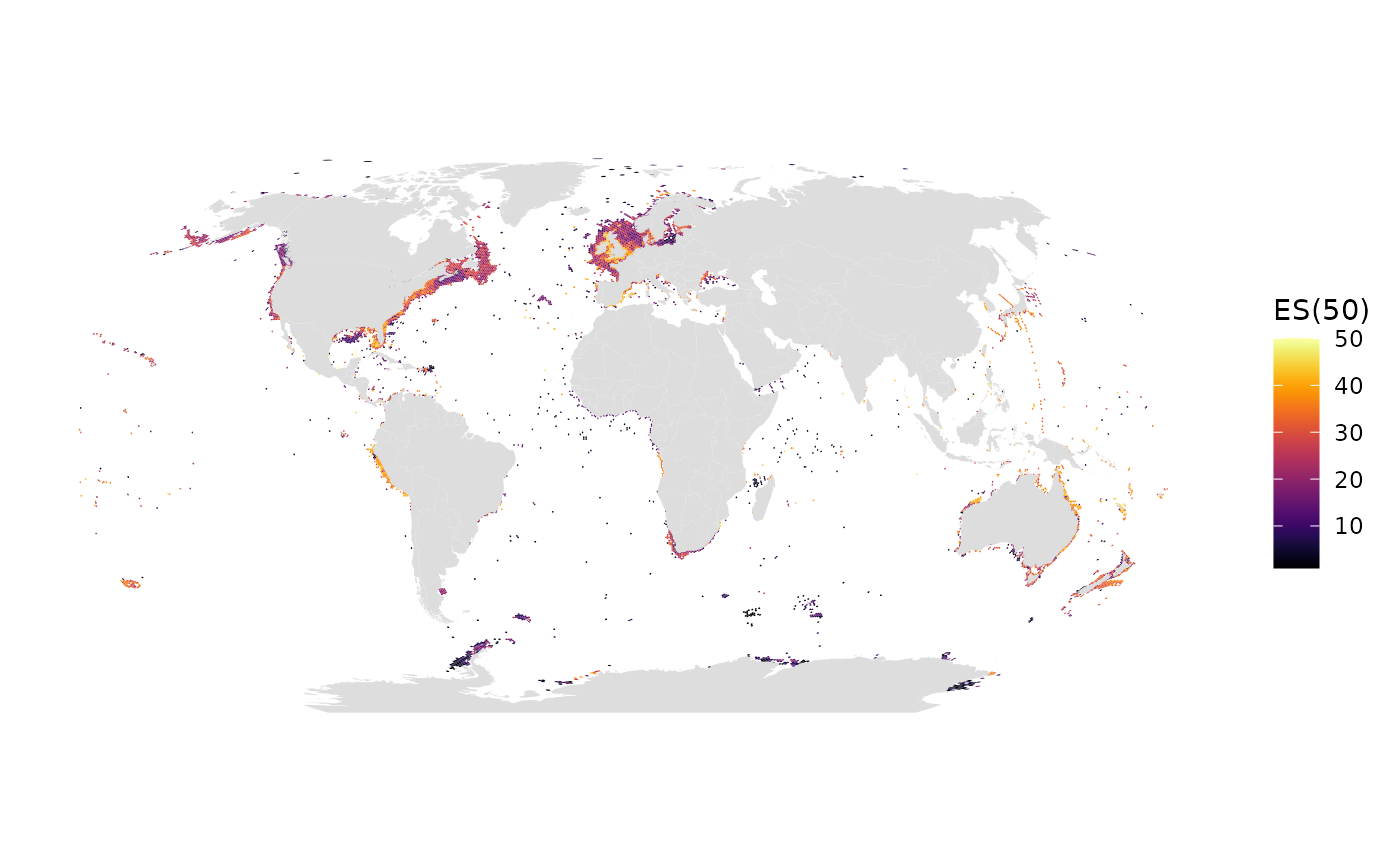
# Shannon index
gmap_indicator(grid, "shannon", label = "Shannon index")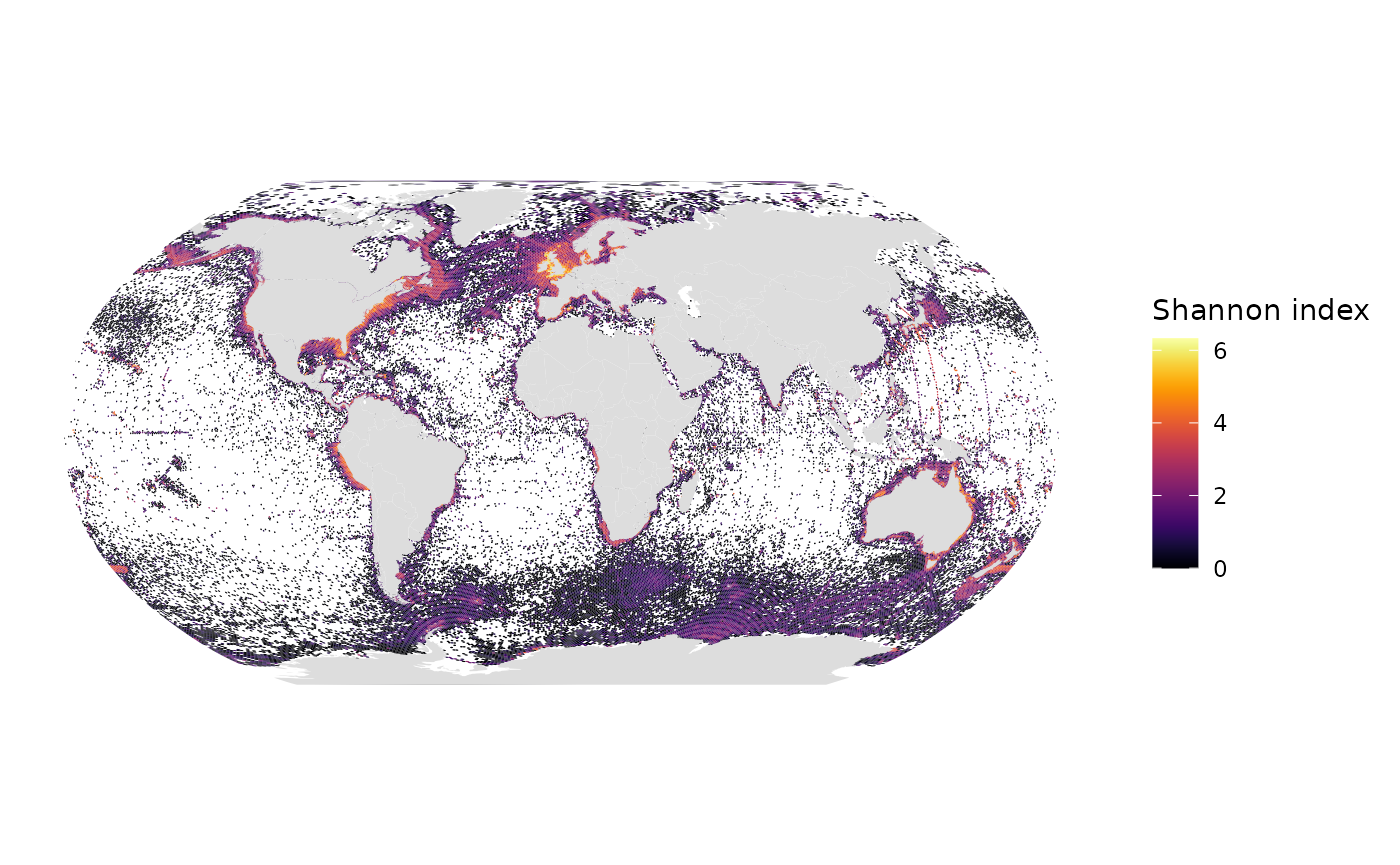
# Number of records, log10 scale, Robinson projection (default)
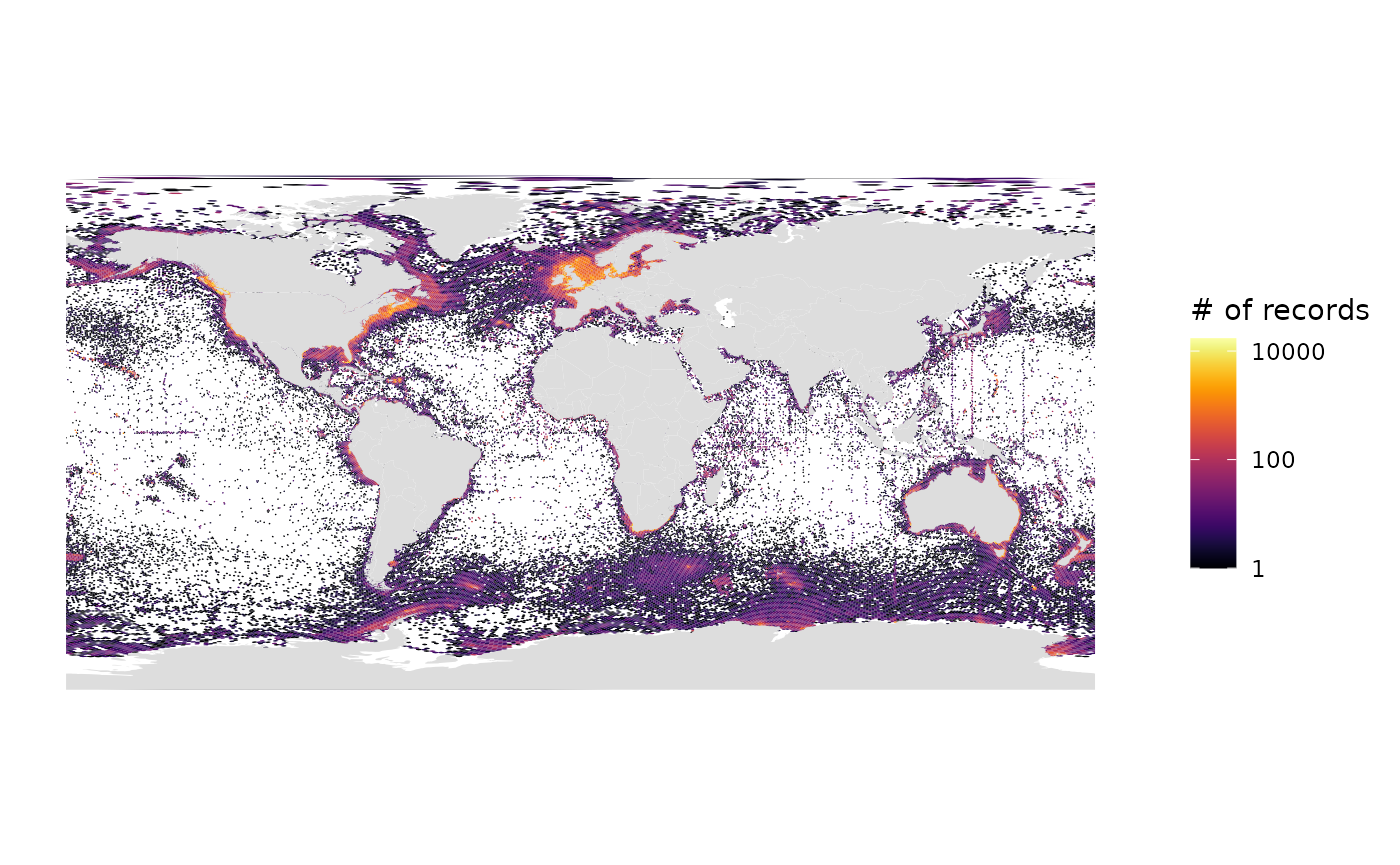
gmap_indicator(grid, "n", label = "# of records", trans = "log10")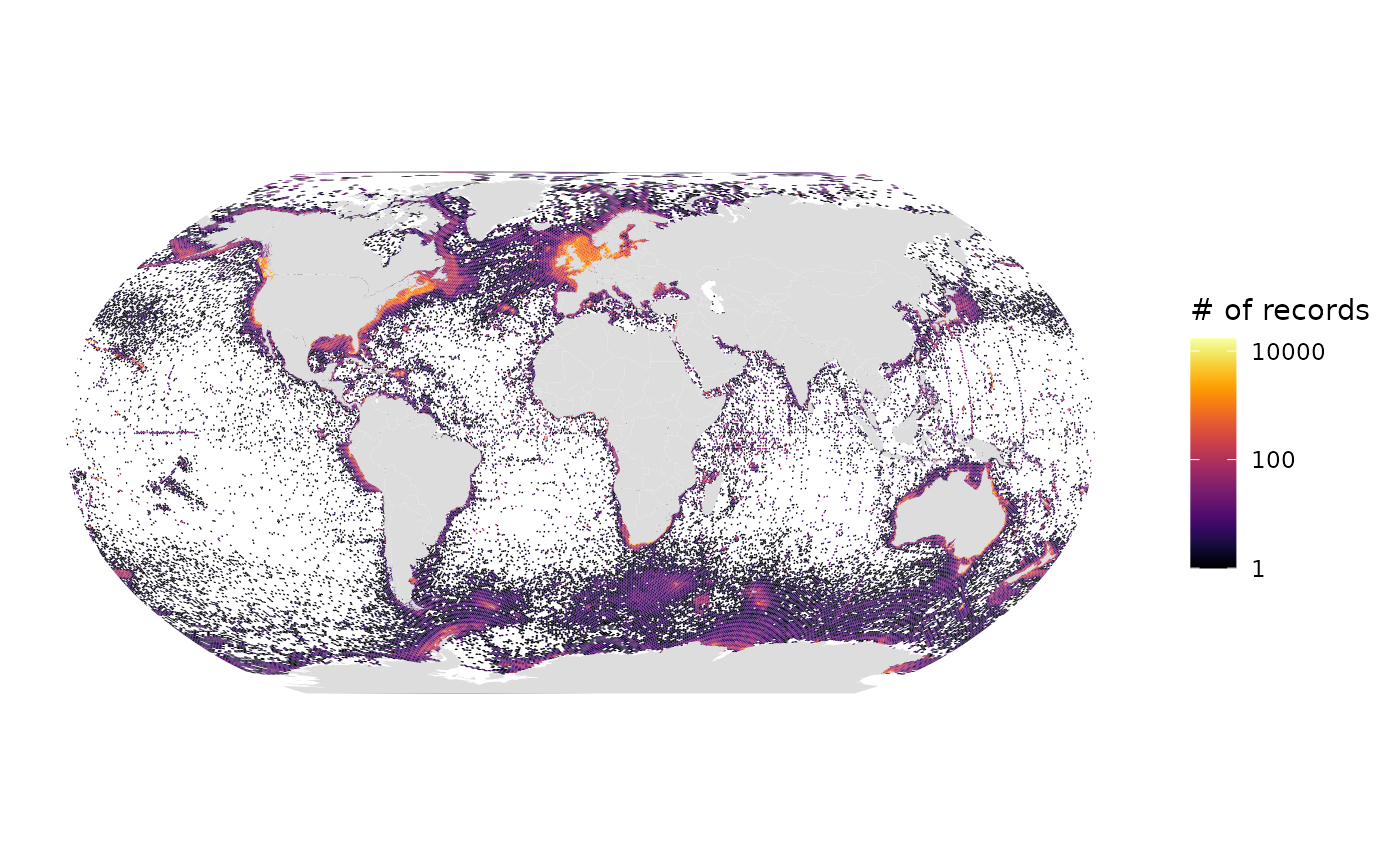
# Number of records, log10 scale, Geographic projection
gmap_indicator(grid, "n", label = "# of records", trans = "log10", crs=4326)