Based on demo script here as presented by Pieter Provoost @ Ocean Data Management for Researchers 2018
# requires devtools::install_github("iobis/robis", ref="obis2")
# robis2 expects these to be loaded (?):
library(ggplot2)
library(tidyverse)
## ── Attaching packages ────────────────────────────────── tidyverse 1.2.1 ──
## ✔ tibble 2.1.3 ✔ purrr 0.3.2
## ✔ tidyr 0.8.3 ✔ dplyr 0.8.3
## ✔ readr 1.3.1 ✔ stringr 1.4.0
## ✔ tibble 2.1.3 ✔ forcats 0.4.0
## ── Conflicts ───────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
sf <- robis::occurrence("Spio filicornis")
## Retrieved 5000 records of 12891 (38%) Retrieved 10000 records of 12891
## (77%) Retrieved 12891 records of 12891 (100%)
dim(sf)
## [1] 12891 120
#robis::map_ggplot(sf)
# TODO: how to inject output of map_leaflet() file here?
# by default this opens in browser at
# file:///tmp/RtmpY77nB4/viewhtml5e25d55bb8f/index.html
str(sf)
## 'data.frame': 12891 obs. of 120 variables:
## $ date_year : int 2010 2001 1997 1996 1973 1991 2010 1979 1982 2000 ...
## $ scientificNameID : chr "urn:lsid:marinespecies.org:taxname:131183" "urn:lsid:marinespecies.org:taxname:131183" "urn:lsid:marinespecies.org:taxname:131183" "urn:lsid:marinespecies.org:taxname:131183" ...
## $ year : chr "2010" "2001" "1997" "1996" ...
## $ scientificName : chr "Spio filicornis" "Spio filicornis" "Spio filicornis" "Spio filicornis" ...
## $ dropped : logi FALSE FALSE FALSE FALSE FALSE FALSE ...
## $ aphiaID : int 131183 131183 131183 131183 131183 131183 131183 131183 131183 131183 ...
## $ decimalLatitude : num 51.3 52.4 54.1 52.8 40.4 ...
## $ language : chr "en" "en" "en" NA ...
## $ subclassid : int 754175 754175 754175 754175 754175 754175 754175 754175 754175 754175 ...
## $ type : chr "sample" "sample" "Sample" NA ...
## $ phylumid : int 882 882 882 882 882 882 882 882 882 882 ...
## $ familyid : int 913 913 913 913 913 913 913 913 913 913 ...
## $ catalogNumber : chr "RSMP_AA_OWF_37_208833" "RSMP_SASBEN0801_16_208047" "RWS1997_OESTGDN14 _ Spio filicornis" "17079" ...
## $ occurrenceStatus : chr "present" "present" "present" "present" ...
## $ terrestrial : logi FALSE FALSE FALSE FALSE FALSE FALSE ...
## $ basisOfRecord : chr "Occurrence" "Occurrence" "Occurrence" "Occurrence" ...
## $ maximumDepthInMeters : num 48 44.5 NA 10.4 29 4 0 NA NA 25.9 ...
## $ modified : chr "2018-04-09 15:18:52" "2018-04-09 15:18:52" "2018-08-07 16:03:26" "2006-06-01 00:00:00" ...
## $ id : chr "0006510d-898e-470c-ad18-aa2cf855d3d7" "0008087f-f8b4-49c0-80fd-0a8d4b38ed22" "000cf7b4-0a3e-407b-be8f-3ed743d5b6a3" "00130ffb-2845-4179-891f-b85a0359bfbb" ...
## $ day : chr "15" "11" "8" NA ...
## $ order : chr "Spionida" "Spionida" "Spionida" "Spionida" ...
## $ dataset_id : chr "a9a3bdc6-209f-4c66-aafd-ce5271cb63b3" "a9a3bdc6-209f-4c66-aafd-ce5271cb63b3" "90411e40-55ce-4ab8-b0d8-29359243e3ce" "386ac1f4-922d-4991-ada2-0ce50dfd3ba7" ...
## $ decimalLongitude : num -4.47 2.11 2.86 4.67 -73.77 ...
## $ date_end : num 1289779200000 997488000000 860457600000 851990400000 120182400000 ...
## $ speciesid : int 131183 131183 131183 131183 131183 131183 131183 131183 131183 131183 ...
## $ occurrenceID : chr "RSMP_AA_OWF_37_208833" "RSMP_SASBEN0801_16_208047" "RWS1997_OESTGDN14 _ Spio filicornis" "urn:catalog:ZMA:Northsea observations:17079" ...
## $ suborderid : int 909 909 909 909 909 909 909 909 909 909 ...
## $ date_start : num 1289779200000 997488000000 860457600000 820454400000 120182400000 ...
## $ footprintSRS : chr "EPSG:4326" "EPSG:4326" "EPSG:4326" NA ...
## $ month : chr "11" "8" "4" NA ...
## $ genus : chr "Spio" "Spio" "Spio" "Spio" ...
## $ eventDate : chr "2010-11-15" "2001-08-11" "1997-04-08T18:25:00+02:00" "1996" ...
## $ eventID : chr "AA_OWF_37" "SASBEN0801_16" "RWS1997_OESTGDN14" NA ...
## $ brackish : logi FALSE FALSE FALSE FALSE FALSE FALSE ...
## $ absence : logi FALSE FALSE FALSE FALSE FALSE FALSE ...
## $ genusid : int 129625 129625 129625 129625 129625 129625 129625 129625 129625 129625 ...
## $ originalScientificName : chr "Spio filicornis" "Spio filicornis" "Spio filicornis" "Spio filicornis (Müller, 1766)" ...
## $ marine : logi TRUE TRUE TRUE TRUE TRUE TRUE ...
## $ minimumDepthInMeters : num 48 44.5 NA 10.4 29 4 0 NA NA 25.9 ...
## $ infraclassid : int 154974 154974 154974 154974 154974 154974 154974 154974 154974 154974 ...
## $ institutionCode : chr "CEFAS" "CEFAS" NA "ZMA" ...
## $ date_mid : num 1289779200000 997488000000 860457600000 836179200000 120182400000 ...
## $ infraclass : chr "Canalipalpata" "Canalipalpata" "Canalipalpata" "Canalipalpata" ...
## $ class : chr "Polychaeta" "Polychaeta" "Polychaeta" "Polychaeta" ...
## $ suborder : chr "Spioniformia" "Spioniformia" "Spioniformia" "Spioniformia" ...
## $ orderid : int 889 889 889 889 889 889 889 889 889 889 ...
## $ datasetName : chr "RSMP Baseline Dataset" "RSMP Baseline Dataset" "Dutch long term monitoring of macrobenthos in the Dutch Continental Economical Zone of the North Sea" NA ...
## $ geodeticDatum : chr "EPSG:4326" "EPSG:4326" "EPSG:4326" "EPSG:4326" ...
## $ kingdom : chr "Animalia" "Animalia" "Animalia" "Animalia" ...
## $ classid : int 883 883 883 883 883 883 883 883 883 883 ...
## $ phylum : chr "Annelida" "Annelida" "Annelida" "Annelida" ...
## $ species : chr "Spio filicornis" "Spio filicornis" "Spio filicornis" "Spio filicornis" ...
## $ subclass : chr "Sedentaria" "Sedentaria" "Sedentaria" "Sedentaria" ...
## $ datasetID : chr "IMIS:dasid:5922" "IMIS:dasid:5922" "IMIS:dasid:5759" "IMIS:dasid:1037" ...
## $ family : chr "Spionidae" "Spionidae" "Spionidae" "Spionidae" ...
## $ kingdomid : int 2 2 2 2 2 2 2 2 2 2 ...
## $ node_id :List of 12891
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "310922b4-9d0c-4de1-92d7-9b442d34765b"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "310922b4-9d0c-4de1-92d7-9b442d34765b"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "310922b4-9d0c-4de1-92d7-9b442d34765b"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "b7c47783-a020-4173-b390-7b57c4fa1426"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "310922b4-9d0c-4de1-92d7-9b442d34765b"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "310922b4-9d0c-4de1-92d7-9b442d34765b"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "310922b4-9d0c-4de1-92d7-9b442d34765b"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "310922b4-9d0c-4de1-92d7-9b442d34765b"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "310922b4-9d0c-4de1-92d7-9b442d34765b"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "b7c47783-a020-4173-b390-7b57c4fa1426"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc" "f92d5d7f-47a6-4605-9fd0-a8538dfde3fd"
## ..$ : chr "1ad35eb9-c615-4733-864a-b585aebcfb70"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## ..$ : chr "4bf79a01-65a9-4db6-b37b-18434f26ddfc"
## .. [list output truncated]
## $ locality : chr NA NA "KLAVERBANK_OV" NA ...
## $ collectionCode : chr NA NA "RWS-boxcore historie" "Northsea observations" ...
## $ dateIdentified : chr NA NA "1997-04-08" NA ...
## $ eventTime : chr NA NA "18:25:00+02:00" NA ...
## $ footprintWKT : chr NA NA "POINT (2.86276 54.082588)" NA ...
## $ country : chr NA NA NA "Netherlands" ...
## $ individualCount : chr NA NA NA "248.7" ...
## $ occurrenceRemarks : chr NA NA NA "individual count in N/m2; location label: 96-coa4" ...
## $ dynamicProperties : chr NA NA NA NA ...
## $ fieldNumber : chr NA NA NA NA ...
## $ bibliographicCitation : chr NA NA NA NA ...
## $ waterBody : chr NA NA NA NA ...
## $ samplingProtocol : chr NA NA NA NA ...
## $ samplingEffort : chr NA NA NA NA ...
## $ sex : chr NA NA NA NA ...
## $ lifeStage : chr NA NA NA NA ...
## $ references : chr NA NA NA NA ...
## $ continent : chr NA NA NA NA ...
## $ scientificNameAuthorship : chr NA NA NA NA ...
## $ specificEpithet : chr NA NA NA NA ...
## $ recordedBy : chr NA NA NA NA ...
## $ coordinateUncertaintyInMeters: chr NA NA NA NA ...
## $ preparations : chr NA NA NA NA ...
## $ taxonRemarks : chr NA NA NA NA ...
## $ materialSampleID : chr NA NA NA NA ...
## $ ownerInstitutionCode : chr NA NA NA NA ...
## $ taxonRank : chr NA NA NA NA ...
## $ identifiedBy : chr NA NA NA NA ...
## $ recordNumber : chr NA NA NA NA ...
## $ otherCatalogNumbers : chr NA NA NA NA ...
## $ eventRemarks : chr NA NA NA NA ...
## $ locationRemarks : chr NA NA NA NA ...
## $ locationID : chr NA NA NA NA ...
## $ county : chr NA NA NA NA ...
## $ stateProvince : chr NA NA NA NA ...
## $ rightsHolder : chr NA NA NA NA ...
## $ institutionID : chr NA NA NA NA ...
## $ license : chr NA NA NA NA ...
## $ coordinatePrecision : chr NA NA NA NA ...
## $ locationAccordingTo : chr NA NA NA NA ...
## $ rights : chr NA NA NA NA ...
## $ associatedReferences : chr NA NA NA NA ...
## [list output truncated]
names(sf)
## [1] "date_year" "scientificNameID"
## [3] "year" "scientificName"
## [5] "dropped" "aphiaID"
## [7] "decimalLatitude" "language"
## [9] "subclassid" "type"
## [11] "phylumid" "familyid"
## [13] "catalogNumber" "occurrenceStatus"
## [15] "terrestrial" "basisOfRecord"
## [17] "maximumDepthInMeters" "modified"
## [19] "id" "day"
## [21] "order" "dataset_id"
## [23] "decimalLongitude" "date_end"
## [25] "speciesid" "occurrenceID"
## [27] "suborderid" "date_start"
## [29] "footprintSRS" "month"
## [31] "genus" "eventDate"
## [33] "eventID" "brackish"
## [35] "absence" "genusid"
## [37] "originalScientificName" "marine"
## [39] "minimumDepthInMeters" "infraclassid"
## [41] "institutionCode" "date_mid"
## [43] "infraclass" "class"
## [45] "suborder" "orderid"
## [47] "datasetName" "geodeticDatum"
## [49] "kingdom" "classid"
## [51] "phylum" "species"
## [53] "subclass" "datasetID"
## [55] "family" "kingdomid"
## [57] "node_id" "locality"
## [59] "collectionCode" "dateIdentified"
## [61] "eventTime" "footprintWKT"
## [63] "country" "individualCount"
## [65] "occurrenceRemarks" "dynamicProperties"
## [67] "fieldNumber" "bibliographicCitation"
## [69] "waterBody" "samplingProtocol"
## [71] "samplingEffort" "sex"
## [73] "lifeStage" "references"
## [75] "continent" "scientificNameAuthorship"
## [77] "specificEpithet" "recordedBy"
## [79] "coordinateUncertaintyInMeters" "preparations"
## [81] "taxonRemarks" "materialSampleID"
## [83] "ownerInstitutionCode" "taxonRank"
## [85] "identifiedBy" "recordNumber"
## [87] "otherCatalogNumbers" "eventRemarks"
## [89] "locationRemarks" "locationID"
## [91] "county" "stateProvince"
## [93] "rightsHolder" "institutionID"
## [95] "license" "coordinatePrecision"
## [97] "locationAccordingTo" "rights"
## [99] "associatedReferences" "verbatimCoordinates"
## [101] "habitat" "organismQuantity"
## [103] "georeferencedDate" "verbatimEventDate"
## [105] "sampleSizeUnit" "georeferencedBy"
## [107] "verbatimLocality" "organismQuantityType"
## [109] "verbatimDepth" "sampleSizeValue"
## [111] "verbatimLatitude" "countryCode"
## [113] "verbatimLongitude" "georeferenceProtocol"
## [115] "georeferenceSources" "parentEventID"
## [117] "collectionID" "nomenclaturalCode"
## [119] "georeferenceRemarks" "dataGeneralizations"
#View(sf)
# TODO: how to inject output of View() here?
table(sf$originalScientificName)
##
## Spio cf. filicornis Spio filicomis
## 6 14
## Spio filicornis Spio filicornis (Mnller, 1776)
## 12164 2
## Spio filicornis (Müller, 1766) Spio filicornis 2
## 597 3
## Spio filicornis agg Spio filicornis aggregate
## 31 36
## Spio filicornis Type A Spio filicprnis
## 7 17
## Spio_filicornis Spiofilicornis
## 12 2
library("magrittr")
##
## Attaching package: 'magrittr'
## The following object is masked from 'package:purrr':
##
## set_names
## The following object is masked from 'package:tidyr':
##
## extract
spio <- robis::occurrence("Spio")
## Retrieved 5000 records of 33712 (14%)
## Retrieved 10000 records of 33712 (29%) Retrieved 15000 records of 33712
## (44%) Retrieved 20000 records of 33712 (59%) Retrieved 25000 records of
## 33712 (74%) Retrieved 30000 records of 33712 (88%) Retrieved 33712 records
## of 33712 (100%)
spio %>% dplyr::group_by(species)%>% dplyr::summarize(n(), mean(decimalLatitude))
## # A tibble: 27 x 3
## species `n()` `mean(decimalLatitude)`
## <chr> <int> <dbl>
## 1 Spio aequalis 16 -39.7
## 2 Spio armata 2917 53.3
## 3 Spio arndti 20 51.6
## 4 Spio bengalensis 2 21.5
## 5 Spio blakei 282 -29.2
## 6 Spio butleri 23 42.7
## 7 Spio cirrifera 23 48.7
## 8 Spio decorata 4651 53.7
## 9 Spio filicornis 12891 53.4
## 10 Spio goniocephala 1750 53.8
## # … with 17 more rows
ggplot2::ggplot(spio) + ggplot2::geom_point(ggplot2::aes(x=decimalLongitude, y=decimalLatitude))
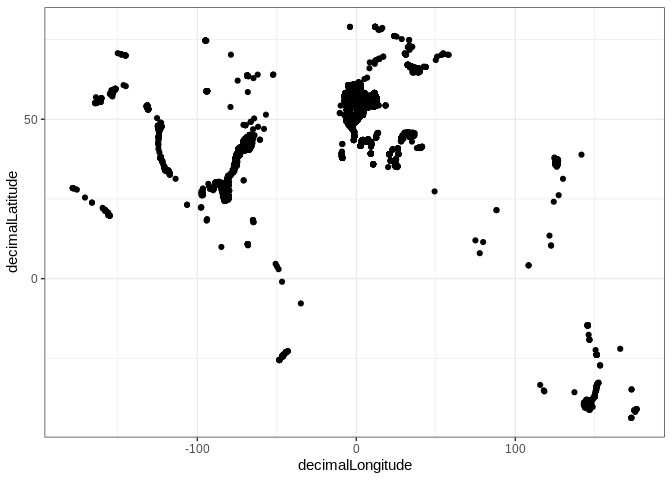
# select one species
#spio %>% dplyr::filter(species == "Spio blakei") %>% robis::map_ggplot()
# === spatial query
# wkt geometry string creation tool: http://iobis.org/maptool/
mol <- robis::occurrence(
"Mollusca",
geometry = "POLYGON ((2.54333 51.07247, 2.10388 51.64189, 2.79053 51.80522, 3.36731 51.36149, 2.54333 51.07247))"
)
## Retrieved 5000 records of 16630 (30%) Retrieved 10000 records of 16630
## (60%) Retrieved 15000 records of 16630 (90%) Retrieved 16630 records of
## 16630 (100%)
#robis::map_ggplot(mol)
ggplot2::ggplot(mol) + ggplot2::geom_bar(ggplot2::aes(date_year), width = 1)
## Warning: Removed 632 rows containing non-finite values (stat_count).
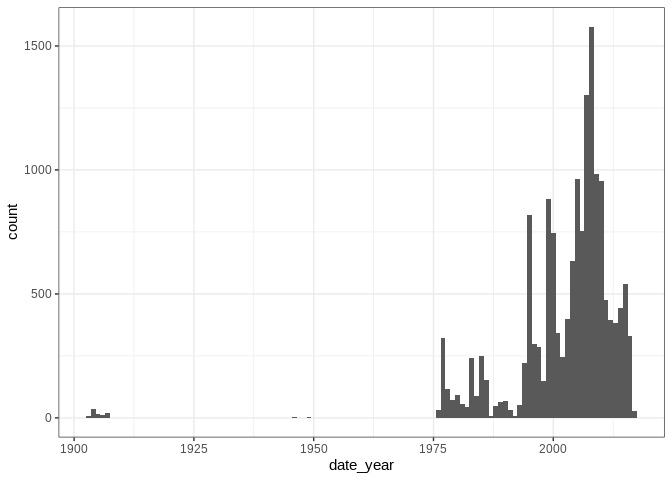
ggplot2::ggplot(mol) + ggplot2::geom_bar(
ggplot2::aes(date_year, fill = class), width = 1
) + ggplot2::scale_fill_brewer(palette = "Spectral")
## Warning: Removed 632 rows containing non-finite values (stat_count).
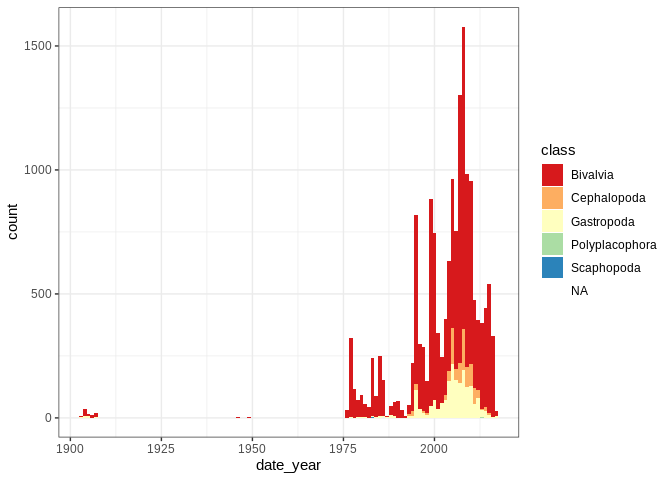
library(marmap)
## Registered S3 methods overwritten by 'adehabitatMA':
## method from
## print.SpatialPixelsDataFrame sp
## print.SpatialPixels sp
##
## Attaching package: 'marmap'
## The following object is masked from 'package:grDevices':
##
## as.raster
library(plotly)
##
## Attaching package: 'plotly'
## The following object is masked from 'package:ggplot2':
##
## last_plot
## The following object is masked from 'package:stats':
##
## filter
## The following object is masked from 'package:graphics':
##
## layout
library(robis)
res <- 0.2
xmin <- 158
xmax <- 180
ymin <- -55
ymax <- -30
nz <- getNOAA.bathy(lon1 = 158, lon2 = 180, lat1 = -55, lat2 = -30, resolution = res * 60)
## Querying NOAA database ...
## This may take seconds to minutes, depending on grid size
## Building bathy matrix ...
nz <- t(nz)
x <- seq(xmin + res / 2, xmax - res / 2, by = res)
y <- seq(ymin + res / 2, ymax - res / 2, by = res)
geom <- sprintf("POLYGON ((%s %s, %s %s, %s %s, %s %s, %s %s))", xmin, ymax, xmin, ymin, xmax, ymin, xmax, ymax, xmin, ymax)
ha <- occurrence("Hoplostethus atlanticus", geometry = geom)
## Retrieved 5000 records of 6631 (75%)
## Retrieved 6631 records of 6631 (100%)
pc <- occurrence("Parapercis colias", geometry = geom)
## Retrieved 5000 records of 23766 (21%) Retrieved 10000 records of 23766
## (42%) Retrieved 15000 records of 23766 (63%) Retrieved 20000 records of
## 23766 (84%) Retrieved 23766 records of 23766 (100%)
plot_ly(z = ~nz, x = ~x, y = ~y) %>%
add_surface(showscale = FALSE) %>%
add_trace(data = ha, x = ~decimalLongitude, y = ~decimalLatitude, z = ~-minimumDepthInMeters, marker = list(color = "#ffcc00", size = 3), name = "Hoplostethus atlanticus") %>%
add_trace(data = pc, x = ~decimalLongitude, y = ~decimalLatitude, z = ~-minimumDepthInMeters, marker = list(color = "#ff3399", size = 3), name = "Parapercis colias")
## No trace type specified:
## Based on info supplied, a 'scatter3d' trace seems appropriate.
## Read more about this trace type -> https://plot.ly/r/reference/#scatter3d
## No scatter3d mode specifed:
## Setting the mode to markers
## Read more about this attribute -> https://plot.ly/r/reference/#scatter-mode
## Warning: Ignoring 495 observations
## No trace type specified:
## Based on info supplied, a 'scatter3d' trace seems appropriate.
## Read more about this trace type -> https://plot.ly/r/reference/#scatter3d
## No scatter3d mode specifed:
## Setting the mode to markers
## Read more about this attribute -> https://plot.ly/r/reference/#scatter-mode
## Warning: Ignoring 3721 observations

#write.csv(mol, file = "mollusca.csv", row.names = FALSE)